Hands-on training program
This program is accepting applications from November 15, 2023 at 12:00 p.m. to February 15, 2024. Acceptance results will be notified via email by the end of February.
You can apply through the abstract submission system.
(This means that abstract submission is required for registration.)
If there is an overwhelming number of applicants, grant recipients will be decided by considering factors such as diversity, whether the applicant has obtained a Ph.D. or whether the applicant is a Ph.D. student.
※ We will cover the travel expenses for those who attend the full duration of the Hands-on training.
Hands-on training programs
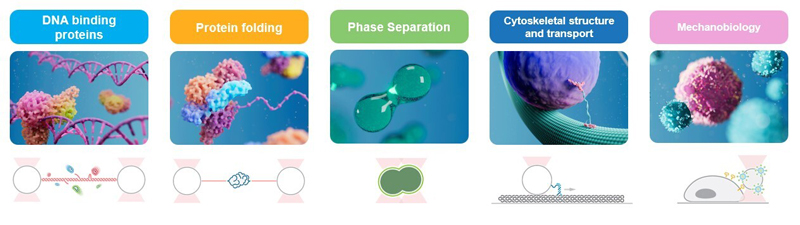
What you learn
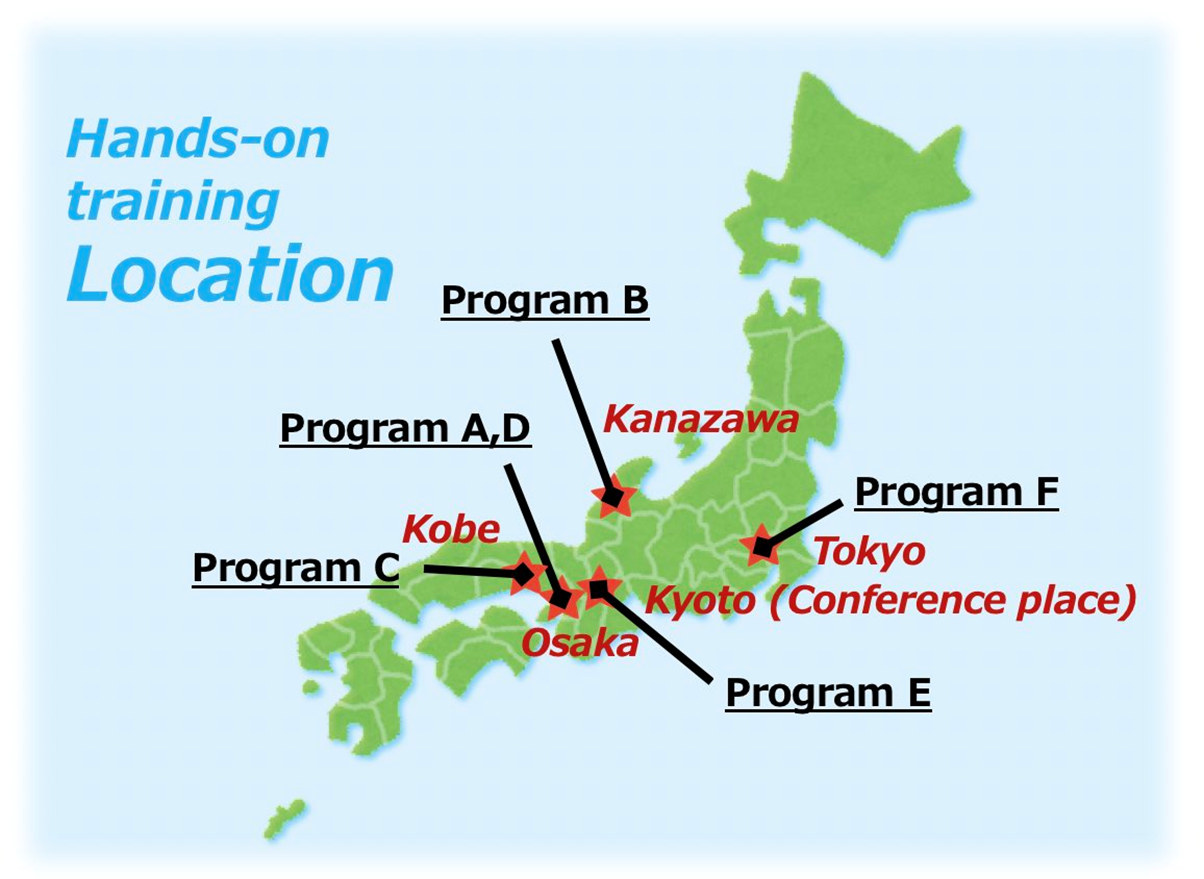
Program A: Millions of single live cell analysis with the automated trans-scale-scope, AMATERAS
Event dates: June 18-21
Venue: Osaka University (June 18-19), RIKEN Kobe Campus (June 20-21)
Participant Capacity: about 6 people
Learning image processing based on information engineering using AMATERAS (a cutting-edge biological microscope, A Multiple/Multiscale Analytical Tool for Every Rare Activity in Singularity) by Osaka University and RIKEN

Program B: Visualizing the nanometer world in liquid by Bio-SPMs
Event dates: July 1
Venue: Kanazawa University
Participant Capacity: about 10 people
One day tour to Kanazawa University to learn about Bio-SPM technologies (super-resolution AFM (FM-AFM & 3D-AFM), high-speed AFM, SICM, AFM for Cell measurement). Participants are given priority to participate in the Bio-SPM summer school conducted annually.
Note: An additional application is required in advance to participate in the Bio-SPM summer school.
- Examples
- - Learning Bio-SPM technologies
- - Tour of SPM imaging of biological molecules or living cells
Program C: CHARMM-GUI/ GENESIS MD tutorial
Event dates: June 30-July 2
Venue: RIKEN Kobe Campus
Participant Capacity: about 20 people
CHARMM-GUI/GENESIS Workshop held in Kobe to learN MD simulations
- Examples
- - Learning GENESIS
- - Learning CHARMM-GUI
- Lecturers
- - Yuji Sugita (RIKEN, Japan)
- - Wonpil Im (Lehign Univ, USA)
Program D: DNA nanomachine tutorial
Event dates: June 22
Venue: Kansai University
Participant Capacity: about 10 people
Molecular robotics is a new academic field that aims to create systems that can be called robots using molecules designed from scratch as components. We are pleased to offer a hands-on program for those interested in molecular robotics to learn the basics of design and evaluation of DNA nanomachines in a short period of time. The program is open to anyone, regardless of specialty or background.
(Sponsored by Grant-in-Aid for Transformative Research Areas (A) “Molecular cybernetics”)
- Examples
- - Learning DNA nanomachine design, visualization, and observation
- Lecturers
- - Masayuki Endo (Kansai Univ, Japan)
- - Akinori Kuzuya (Kansai Univ, Japan)
- - Yuki Suzuki (Mie Univ., Japan)
- - Ibuki Kawamata(Tohoku Univ, Japan)

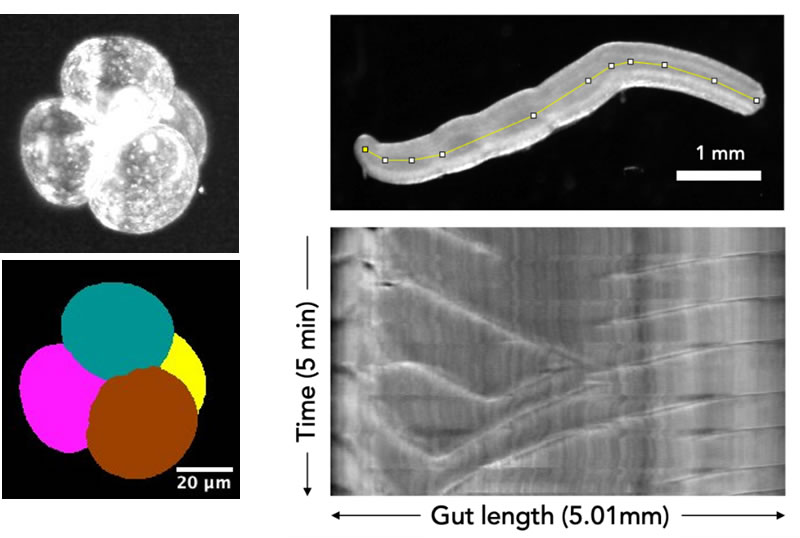
Program E: Exploring multi-cellular mechanics
Event dates: June 29
Venue: Kyoto University
Participant Capacity: about 5 people
This hands-on session will provide an introduction to imaging and image analysis of multi-cellular systems such as tissue and embryo. Participants will learn how to handle these specimens, take images (2D, 3D), and extract biophysical parameters by image analysis.
- Topics
- ・Microscopic observation and image analysis of gut motility.
- ・Handling mouse embryo
- ・3D imaging of mouse embryo by light-sheet microscopy
- ・Live-cell high-speed AFM
- Lecturers
- - Takafumi Ichikawa (Kyoto U.)
- - Masafumi Inaba (Kyoto U.)
- - Shige H. Yoshimura (Kyoto U.)
- Sponsored by:
- Grant-in-Aid for Transformative Research Areas (A)
“Mechanical Self-transformation of Living Systems”
Program F: Real-time single-molecule experiments with optical tweezers
and correlated fluorescence microscopy
Event dates: June 22-23
Venue: Institute for Quantitative Bioscience, The University of Tokyo
Participant Capacity: about 10 people
Witness the seamless integration of optical tweezers and fluorescence microscopy into a single state-of-the-art system: the C-Trap! This one-day workshop in Tokyo includes a brief introduction to the developments and applications of optical tweezers and correlated microscopy in biological research by Institute For Quantitative Biosciences, Tokyo University and LUMICKS. It will be followed by a live demonstration where molecular interactions and protein activity of model systems will be visualized in real-time, at the single-molecule level and with unparalleled resolution. You will also be able to get some hands-on experience with single-molecule assays yourself!